A Computer Network Enabling an Increasing Volume of Data
The National Institute of Genetics (NIG) at the Research Organization of Information and Systems, an inter-university research institution corporation, has built and is now operating an international database of nucleotide sequences called the DNA Data Bank of Japan, or DDBJ, and is offering computing services using its supercomputer. We spoke with two NIG researchers, Yasukazu Nakamura, a professor at the Genome Informatics Laboratory, and Osamu Ogasawara, an assistant professor at the Laboratory for Gene-Expression Analysis, about NIG’s activities and the role played by SINET.
(Interview date: November 14, 2013)
First of all, can you give us an overview of the National Institute of Genetics?
Nakamura:NIG was established in 1949. As a core research institution in the life sciences domain in Japan, it engages in many different studies. As an inter-university research institute, it offers opportunities and facilities for joint studies to researchers across the country. It also serves as the Department of Genetics in the School of Life Science of the Graduate University for Advanced Studies. You could see it as an institute that is comprehensively engaged in advanced studies and education in genetics.
Its specific activities include the National BioResource Project. It develops, collects, and offers bio-resources for academic purposes, such as mice and drosophilae, commonly known as fruit flies. It is building and operating a database of biological genetic resources that is open to the public. Another example is the Advanced Genomics Project, in which genomic data are analyzed using a leading-edge next-generation sequencer. And a third example is the project I am involved in, which is DDBJ, an international database of DNA nucleotide sequence information.
Ogasawara:Soon after the Sanger method of DNA sequencing had been developed, it was expected that a considerable amount of nucleotide sequence data would definitely be created in the future. In response to this, there had been increasing momentum in the West to create an international database, and there was a call for Japan to join this initiative. At the moment, DDBJ is working with GenBank provided by the U.S.-based National Center for Biotechnology Information (NCBI) and the ENA of the European Bioinformatics Institute (EMBL-EBI) to create and operate the International Nucleotide Sequence Database (INSD).
What is the objective and significance of DDBJ’s activities?
Nakamura:Genetic nucleotide sequence data are associated with studies in a wide range of disciplines, including biology, medicine, and agricultural sciences. It is important for research to make clear which nucleotide sequence is related to which phenomenon. Take biological evolution, for example. If a comparison of the nucleotide sequence between two organisms discovers two species with similar sequences, that means that they are close in their evolutionary lineage. If their nucleotide sequences differ significantly, they are distant from each other. However, if these nucleotide sequence data aren’t systematically archived, it will be impossible to reuse the data or to repeat the test. DDBJ uniformly assigns ID codes to the individual nucleotide sequence data stored in it, and organizes the associated data, such as the location where the sample was taken and the part of the organism. That allows researchers to increase the accuracy of their research and efficiency. When they write academic papers related to genes, they are required to specify the ID codes assigned by DDBJ, GenBank, or EMBL-Bank.
We hear that research data are increasing at an accelerating pace in this discipline.
Nakamura:A major reason for this growth is the dramatic improvement in the performance of the sequencers used to determine the nucleotide sequences. Among others, the performance of the latest system, called the next-generation sequencer, is four to five digits higher than the conventional system. Accordingly, the volume of the data archived is growing. In addition, the INSD is designed to ensure that DDBJ, GenBank, and EMBL-Bank share the same data. A massive amount of data are copied between them. The performance level required for another service that NIG provides, specifically its computing service using its supercomputer, is also becoming higher and higher.
It sounds as if the ICT infrastructure that supports research activities is subject to a heavy load. What is its environment like at the moment?
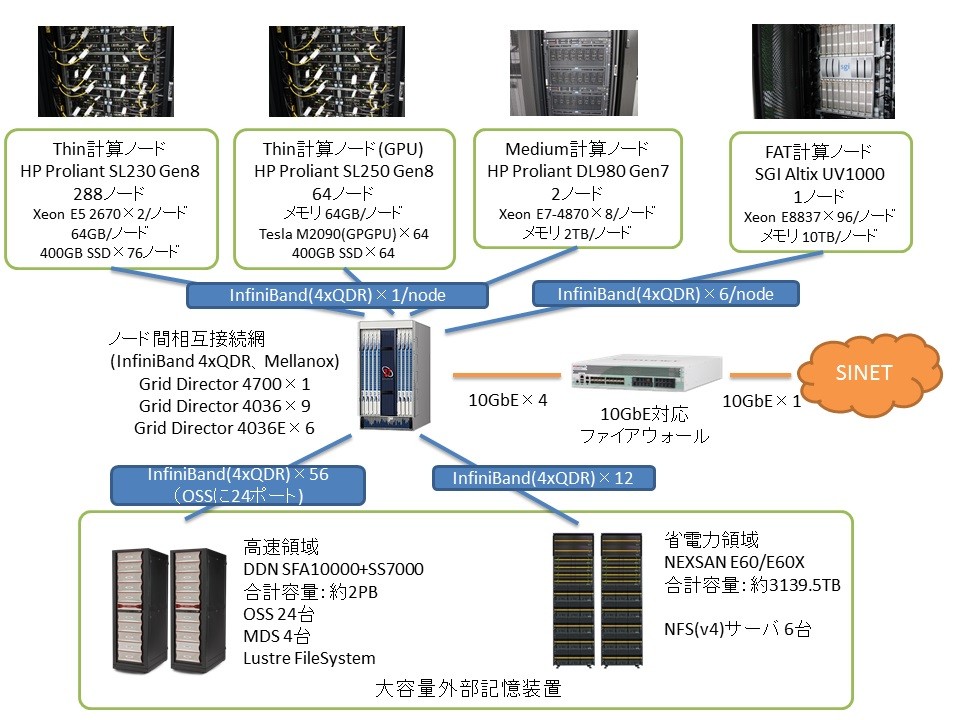
Ogasawara:Regarding the supercomputer, its architecture was revised at the time of the recent procurement, and the operating software was completely rewritten toaccommodate the new architecture.. As a result, its operational performance has been massively improved. The previous supercomputer had a logical peak operation performance of 20 TFLOPS, whereas the current one has reached 169 TFLOPS. This is expected to increase to 366 TFLOPS after an interim upgrade. An upgrade was made to its storage capacity as well. The previous supercomputer had 0.65 PB on disk and 0.75 on tape. The current one has a capacity of 2 PB for computation and a separate capacity of 3 PB for the archive. After the interim upgrade, these will be raised to 7 PB and 5.5 PB, respectively.
A notable characteristic of the system lies in its heavy I/O load. At peak times, there are as many as 500,000 data entries a day. It is fairly difficult to create database indices. We pay a lot of attention to increasing the speed of the I/O process. For example, we have introduced a Lustre parallel-distributed file system. Regarding computer nodes, there is one FAT node with a 10 TB memory, two medium nodes with 2 TB of memory each, and 352 thin nodes with 64 GB of memory each, all of which are in operation.
What about the network?
Ogasawara:Nearly 1,100 researchers have registered as DDBJ users. Almost every day, they upload nucleotide sequence data and conduct analysis using our supercomputer. Aside from that, DDBJ exchanges data with GenBank and EMBL-Bank. The traffic from outsiders is much higher than that inside the NIG premises. The data stored in DDBJ and the supercomputer computing resources constitute the key infrastructure supporting Japan’s DNA research, which is why the network needs to have a high level of performance and reliability. As SINET provides solid support in this respect, NIG manages to provide stable service to researchers. At the moment, we always have a bandwidth of 3 to 4 Gbps, and we have no complaints about the network.
Lastly, can you talk about future developments?
Nakamura:DDBJ will continue to fulfill its key missions of continuing to archive massive nucleotide sequence data and provide computer resources. However, as we have mentioned many times, there is no sign of a halt in the growth of data capacity or in performance improvements in sequencers and computers. As there is demand to make the most of network bandwidth, we have great expectations for the future development of SINET.